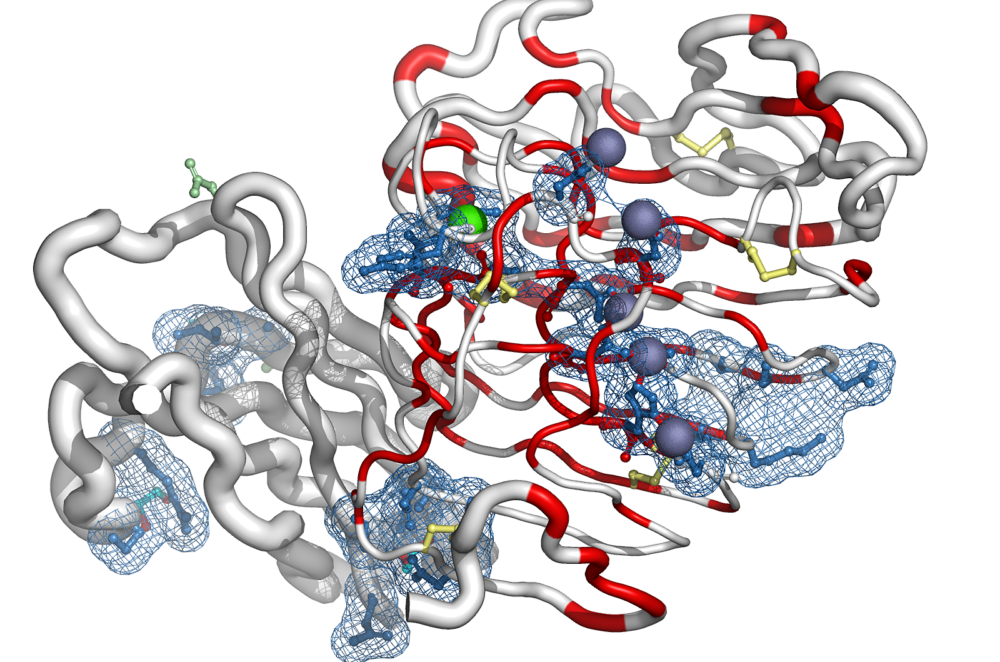
Le professeur Patrice Gouet Biologiste de l’IBCP de Lyon, nous parlera de « Foldscript : a new webserver for interpreting artificial intelligence in structural biology », le jeudi 03 avril à 13H en salle F4.
3D modelling programs based on artificial intelligence (AI) have revolutionised structural biology, often predicting protein structures with an unprecedented level of confidence. However, these structures remain models and it is always interesting to compare the results of different predictors (AlphaFold3, RoseTTAFold2, ESMFold…) and not to limit the choice to the model generated with the best confidence score; it can also be crucial to introduce experimental data, such as known intermolecular regions, in the selection of the best model. To address these issues, we have created a web server called « FoldScript » (https://foldscript.ibcp.fr). It can be used to synthesise the multi-model information generated by the AI in 1D, 2D and 3D. We have tested this web software on numerous viral protein complexes studied by our team, for which low and high confidence scores have been obtained. We show that while the AI can provide relevant answers, a critical view remains crucial. Indeed, the « best » solution proposed by the AI is not always the true « best » solution.